Membrane proteins under investigation
About 30% of the natural occuring proteins in a cell are membrane proteins. As key players in cellular processes these proteins mediate various cellular functions including signal transduction, cell adhesion, transport across the membrane, etc. To understand the role of membrane proteins and how they interact with each other in their natural environment it is of great importance to know the structure of these proteins. Therefore we combine recombinant protein expression, peptide synthesis and spectroscopic methods including nuclear magnetic resonance, circular dichroism and fluorescence spectroscopy to investigate structure, alignment and function of different membrane proteins. Two examples of our work are presented in the following.
Classically, the binding of the ligand to the platelet-derived growth factor receptor β (PDGFR), a receptor tyrosine kinase amogst others involved in cell proliferation, leads to receptor dimerization triggered by the transmembrane segments and thus to trans-autophosphorylation of several tyrosine residues in the cytoplasmic domain of the receptor, which increases the kinase activity of PDGFR and creates binding sites for SH2 domains, whereby downstream signaling is activated.
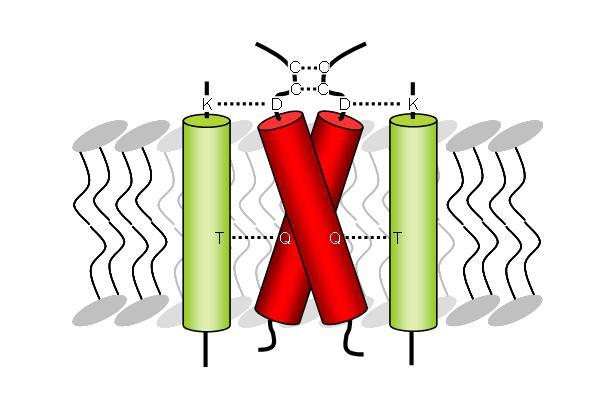
The viral E5 oncoprotein of the bovine papillomavirus type I is a small transmembrane protein consistent of 44 mostly hydrophobic residues. As a disulfide-bridged homodimer E5 mainly interacts with the PDGFR, resulting in cell proliferation independent of the natural ligand. The structural basis for the helix-helix interactions between E5 and the receptor has been attributed to specific residues in the transmembrane and juxtamembrane domains of both proteins (Figure 1). We intend to carry out a PISEMA analysis to investigate the molecular interactions between E5 and the transmembrane domain of the PDGFR, since very little is known about such molecular interfaces occurring within a hydrophobic lipid bilayer. Our investigations will improve the understanding of the E5-mediated activation of the receptor and furthermore will allow us to get insights to manipulate the function of transmembrane receptors using hydrophobic effector molecules.
The twin arginine translocation (Tat) system transports fully folded proteins, including their cofactors, across bacterial and thylakoid membranes. Proteins destined for this pathway are synthesized with an N-terminal signal peptide containing a highly conserved motif with two consecutive arginine residues. The minimal Tat system, which is active in B. subtilis, consists of only two proteins, namely TatAd and TatCd, which are responsible for the translocation of the folded phosphodiesterase (PhoD) into periplasm.
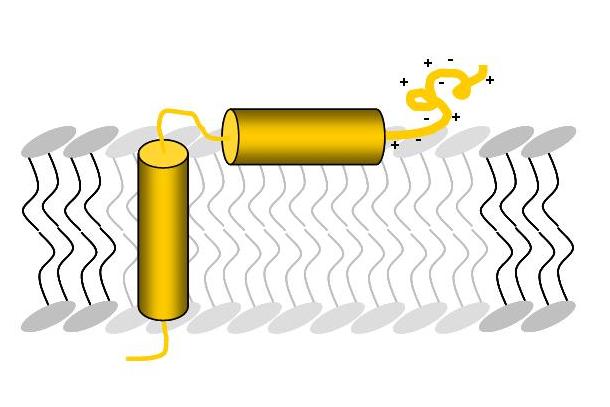
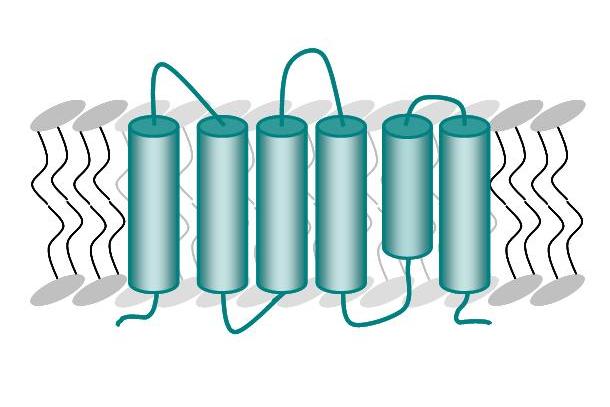
The transmembrane protein TatAd is the pore forming unit of this transport system in B. subtilis and consists of a transmembrane helix, an amphipathic helix attached to the surface of the membrane and a highly charged, unordered C-terminal part (Figure A). The membrane protein TatCd (Figure B) is predicted to contain six transmembrane segments.
In our lab we developed a multi-construct approach to investigate TatAd, where three different parts of the protein were constructed and examined separately, in pairwise combination or as full-length protein. The respective structure of the protein was successfully examined using CD, oriented CD and solution NMR, while the alignment within the membrane was elucidated by solid state NMR. Our NMR results confirm previous studies and provide first detailed structural information about TatAd in a natural environment. We also investigated the conditions of the reconstitution of the TatCd in lipid bilayers and estimated the helicity of the protein in different lipid and detergent systems. Our aim is to characterize the structure and functionality of the Tat translocase, especially by combining the TatCd component as a complex with TatAd and with the signal peptide of PhoD.