NMR spectroscopy
Solid state NMR spectroscopy is our major tool to shed light on the structural basis of the function of membrane-active peptides. Making use of orientation dependence of NMR in uniaxially aligned systems, we are able to access the alignment of peptides in the membrane, but also their conformation and dynamics.
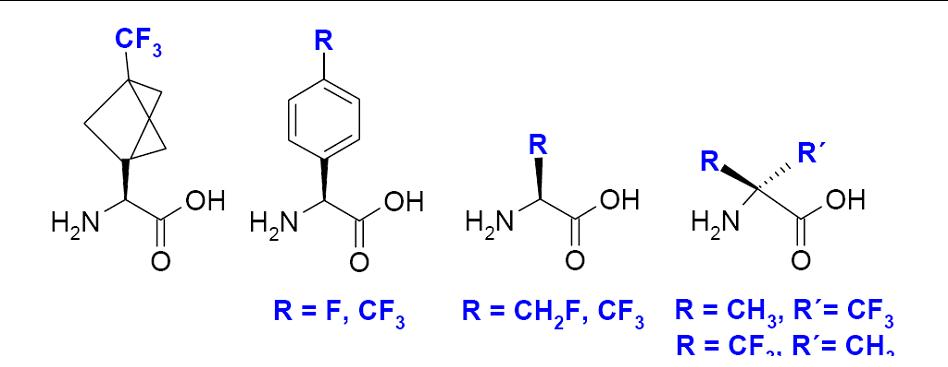
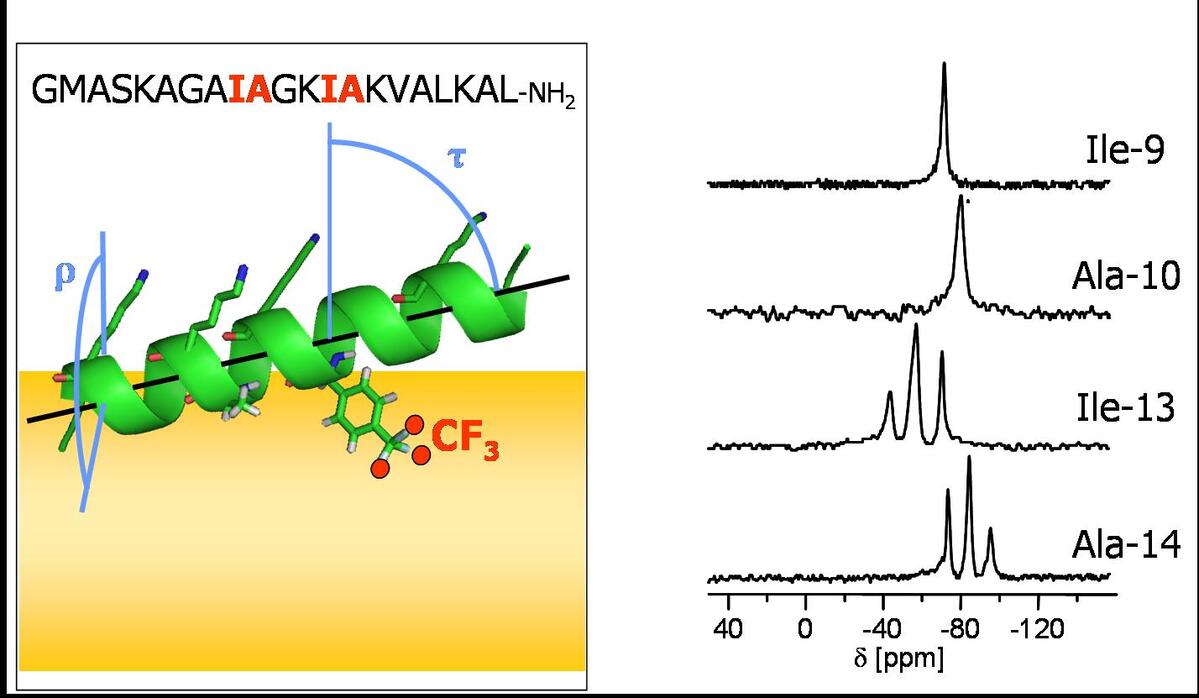
Besides using conventional isotope labels such as 2H and 15N, our group has introduced and extensively applied a highly sensitive 19F-labeling scheme based on the new designer-made amino acids CF3-Bpg, and CF3-Ala. They provide a direct link to the backbone, and measuring the anisotropic 19F-19F dipolar coupling allows then to determine the peptide alignment.
If the secondary structure of a peptide or protein segment is known, its alignment with respect to a lipid bilayer can be determined from the NMR signals of several 2H, 15N or 19F labels. To this aim, a series of peptides, each carrying a single label in a varying amino acid position, is prepared. The sidechain orientation of each peptide is then determined from the orientation dependent quadrupolar coupling (2H) or homonuclear dipolar coupling (19F), which when combined yield the peptide alignment.
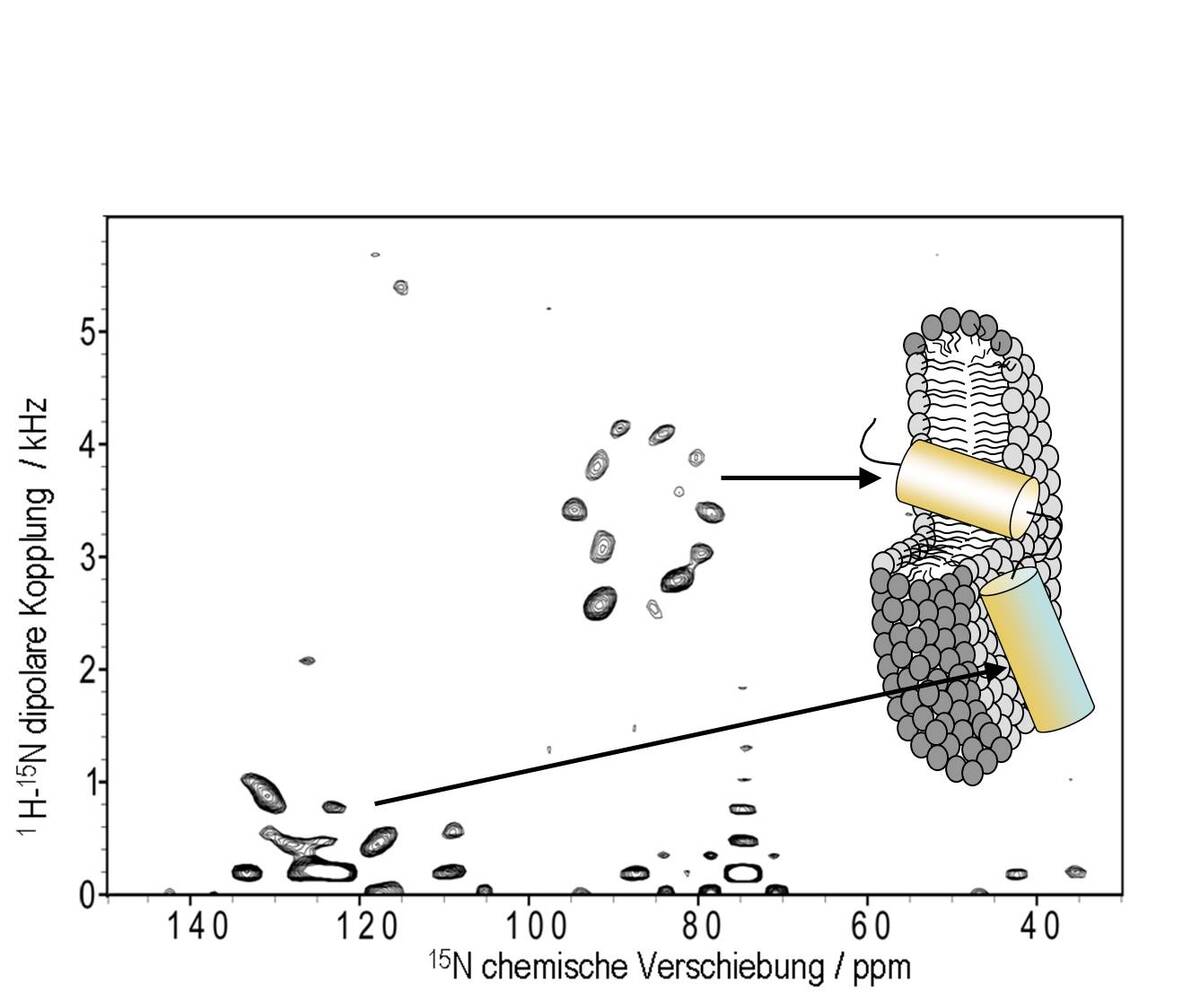
To obtain the structure of small membrane proteins such as TatA we use a combined approach of solution NMR in detergent bicelles and solid state NMR in bicelles or oriented membranes. Here, solution state NMR is able to yield the local structure at high resolution, whilst orientational information from solid state NMR is needed to position different segments correctly with respect to the bilayer and to each other. Such orientational information is obtained from 1H-15N dipolar coupling and the 15N chemical shift anisotropy, determined from uniformly 15N labeled peptides using seperated local field techniques such as the PISEMA experiment.
Peptides are highly mobile in a membrane environment. In such systems NMR monitors averages of structural parameters rather than rigid structures. However, we were able to refine the analysis of the orientation dependent NMR data to access as well the distribution of structural parameters, this way getting insight into the dynamical behavior of membrane active peptides.
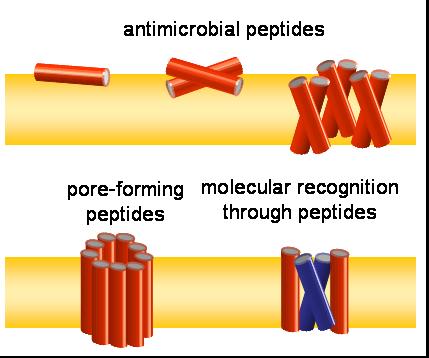
Often, peptides act as oligomers, e.g. as synergistic hetero-dimers like PGLa and magainin 2, or as homo-oligomeric pores such as mellitin. The formation of such assemblies is reflected in the averaging of the NMR signal or changes of peptide alignment, which we use to characterize the different oligomeric states of membrane active peptides. Furthermore, we use solution NMR to map the interface of interacting peptides, and solid state NMR distance measurements to validate oligomer formation in the membrane.
Solid state 19F NMR in biological samples imposes several hardware challanges. For example, the resonance frequencies of 19F and 1H are very close and difficult to isolate, and the often high salt content of biological samples can cause sample heating. To this aim we use dedicated home-built NMR probes with special double-resonance circuit designs and resonators which reduce the electric field component to avoid sample heating.