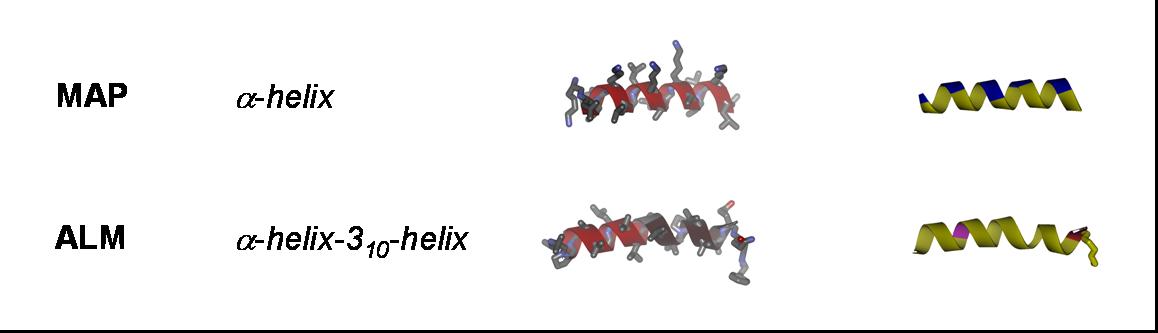Membrane - active peptides under investigation
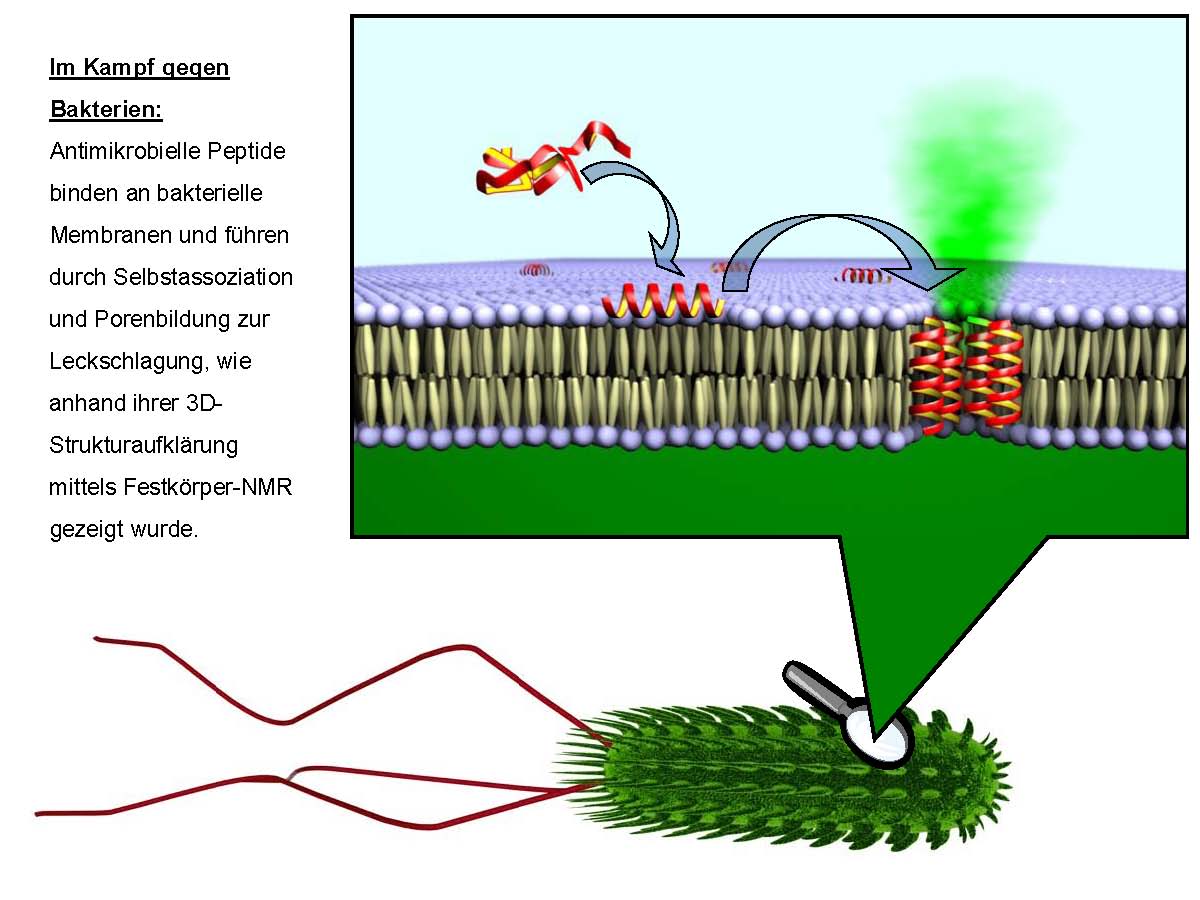
The main focus of the BioNMR group is natural or designer-made peptides interacting with biological membranes. Thus, we study peptides that are uniform in their abilities to bind to the lipid bilayers (i.e. membrane-active peptides) and for which this particular molecular mechanism is functionally important (i.e. membranotropic peptides).
The various systems we are studying manifest different kinds of biological activities, and are commonly classified as: antimicrobial, fusogenic and cell-penetrating peptides, and transmembrane domains of membrane proteins.
We are currently working on the following peptides, which are studied in depth and compared with one another:
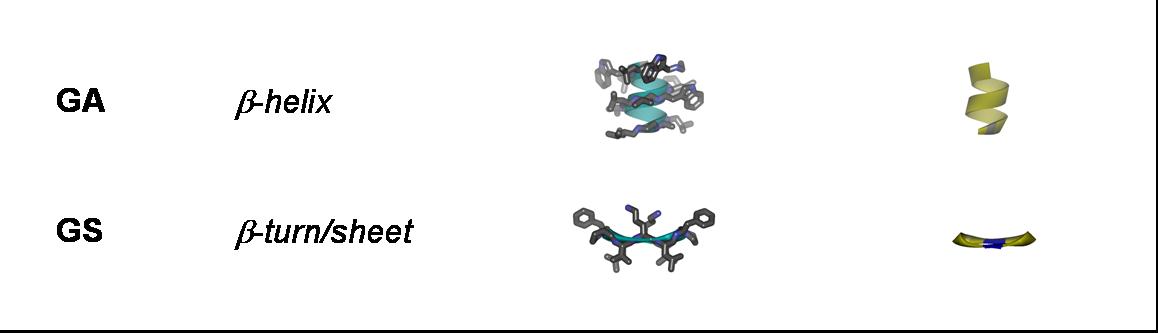
Antimicrobial peptides: Gramicidin S (GS), gramicidin A (GA), PGLa, magainin 2 (MAG2), temporin A (TA), MSI-103, and alamethicin (ALM);
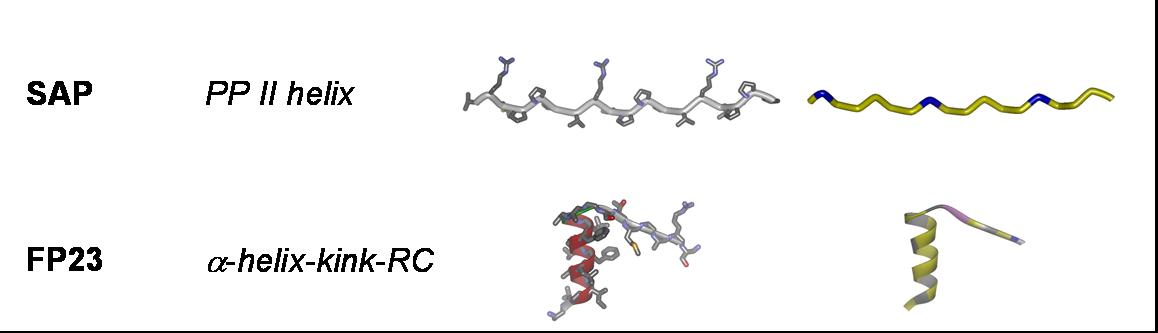
Fusogenic peptides: B18 and FP23;
Cell-penetrating peptides: TAT; R8 ((Arg)8), model amphipathic peptide (MAP), TP10, sweet arrow peptide (SAP), and BP100;
Transmembrane peptides – segments of membrane proteins: WLP23, E5, PDGFR-TM, TCRα-TM, MscL M1-TM, and influenza virus M2-TM;
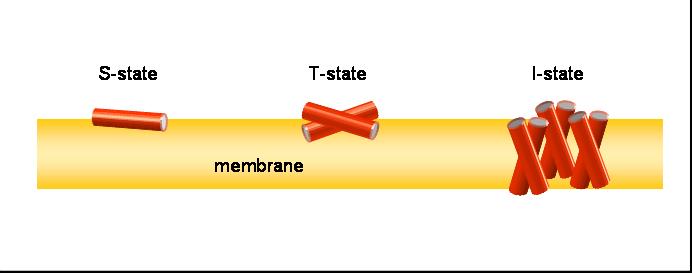
Our aim is to experimentally characterize these peptides in terms of their secondary structure, topology, alignment, aggregation status and dynamics. This allows us to compare the free vs. membrane-bound forms, in order to correlate the above properties with the respective functional mechanisms. Additionally, by cross-comparing structurally variable peptides we accumulate knowledge about general principles of the structure-function relationships in membrane-active peptides. The peptides listed above cover a wide range of different characteristics, allowing such correlations. They differ in: amino acid composition, e.g. rich in arginine (TAT, R8), α-aminoisobutyrate (ALAM), lysine (BP100), or glycine (FP23); in their 3D folding properties, e.g. amphiphilic α-helices (MAP, PGLa, MSI-103, MAG2, BP100) or β-strands (e.g. GS), versus flexible sequences (FP23, TAT); in charge, e.g. TAT has a net charge of +8 while ALAM has a charge of -1, and the related peptides PGLa (+5), MAP (+6), MSI-103 (+7); and in length, e.g. amphiphilic helices from 23 amino acids (MAG2) down to 11 (BP100).
All of these membrane-active molecules present a challenge to conventional structure analysis by X-ray diffraction of crystals or NMR in solution; hence we have developed and applied novel solid-state 19F-NMR techniques in a combination with optical spectroscopy methods to reveal structural information in a biologically relevant context, i.e. in the membrane-bound state under quasi-native conditions.
For 19F-NMR labeling we mostly use in-house chemically synthesized peptide analogues employing novel fluorine-containing amino acids as labels, but also 2H, 15N, or 13C isotopes are used and introduced via standard recombinant expression approaches.