Research
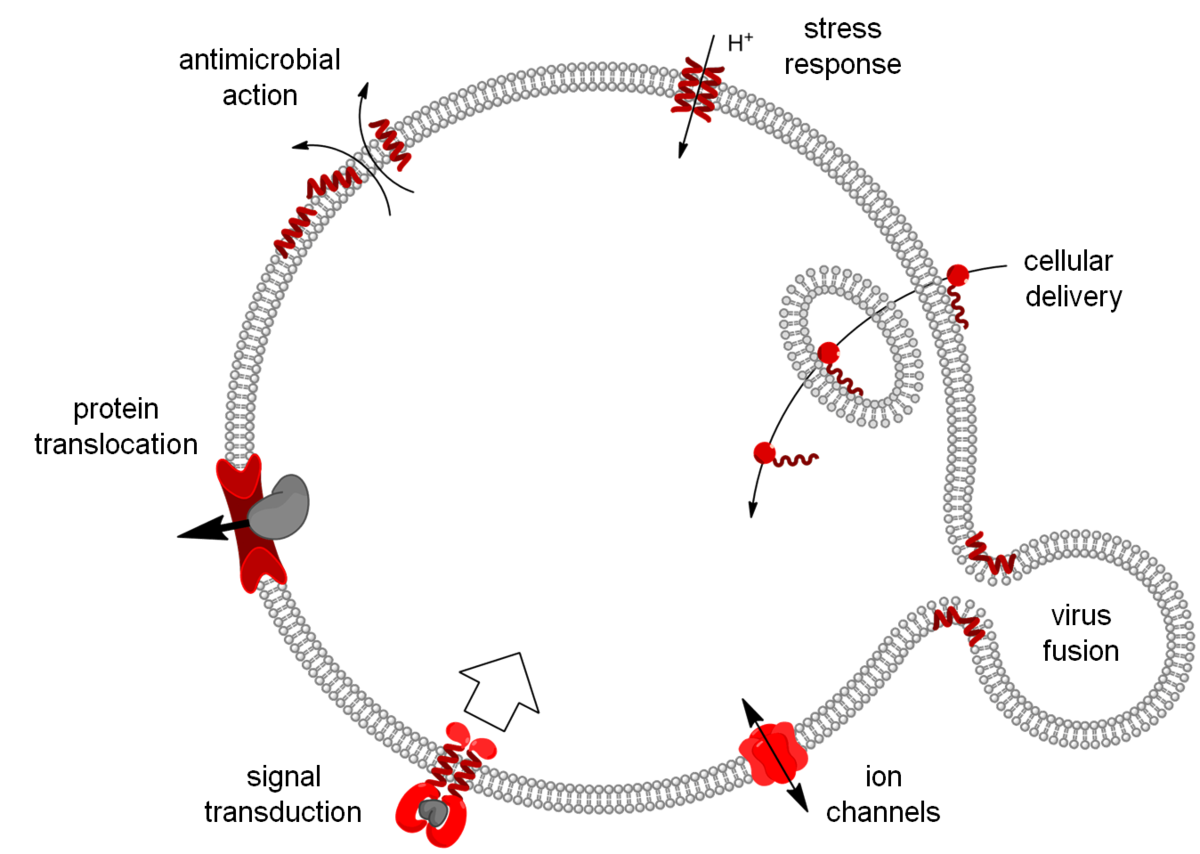
Our research is focused on molecular tools and machineries that are involved in transport across biomembranes. This includes direct bilayer permeabilization by antimicrobial, cell-penetrating and fusogenic peptides, but also specific ion transport by biofilm-inducing peptides and ion channels, as well as highly orchestrated events such as protein translocation and tyrosine-kinase receptor signaling.
Solid-state NMR spectroscopy with selective 19F-labels (and/or amino acid type selective 15N-labels) in aligned samples offers the advantage of studying membrane-bound peptides and transmembrane protein segments in a highly sensitive manner under quasi-native conditions, i.e. embedded in any desired lipid composition and at ambient temperature. This way, dynamical properties and structural re-arrangements can be addressed in a biologically relevant environment, and the lipid molecules and morphological changes of the bilayer can be observed at the same time. Our methodological spectrum is rounded off by other biophysical methods, such as circular dichroism (at our KARA synchrotron-beamline UV-CD12), FRET, DSC, MD simulations (in cooperation), etc., besides the appropriate (micro)biological and functional assays.
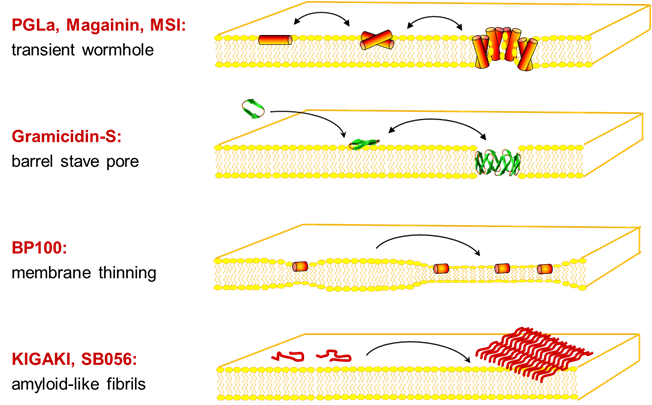
For structure analysis of peptides we designed a series of conformationally constrained 19F-labeled amino acids [1-3], which can be observed even in native biomembranes due to the absence of a natural abundance background [4]. 19F-NMR can thus be used to characterize the conformation, alignment, dynamics and oligomerization of membrane-bound peptides with various structures, e.g. alpha-helices, beta-sheets, intrinsically disordered in the 2D plane, PPII-helices, etc. This way, the molecular mechanisms of several representative peptide antibiotics could be structurally demonstrated at the quasi-atomic level [5-7].
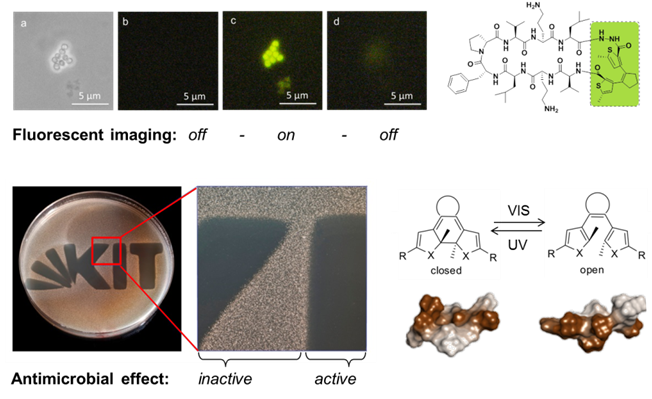
Several designer-made sequences showed an undesirable tendency to aggregate as amyloid-like fibrils in membranes [8]. Even more alarming was the observation that certain antimicrobial peptides triggered bacterial biofilms at sub-inhibitory concentrations, thereby offsetting their advantage of not inducing resistance [9]. These insights were used to design a photo-switchable antibiotic that can be controlled by light [10], and the same concept is now being applied against cancer.
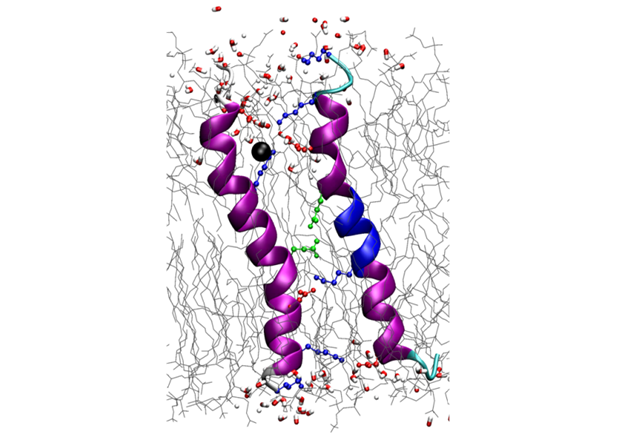
The biofilm-inducing effect of amphiphilic helical antimicrobial peptides could be explained by their structural similarity to a peptide from a toxin/antitoxin system, which short-circuits the electrochemical gradient across the plasma membrane, leading to ATP depletion and dormancy. Structure analysis of this bacterial stress-response peptide TisB revealed a transmembrane antiparallel dimer, held together by four salt bridges and a H-bond [11]. Charge mutations proved that TisB employs this unusual motif of an electrostatic “charge zipper” (see below [12, 13]) to selectively transport protons across the hydrophobic lipid barrier. Other biofilm-modifying peptides are currently under investigation.
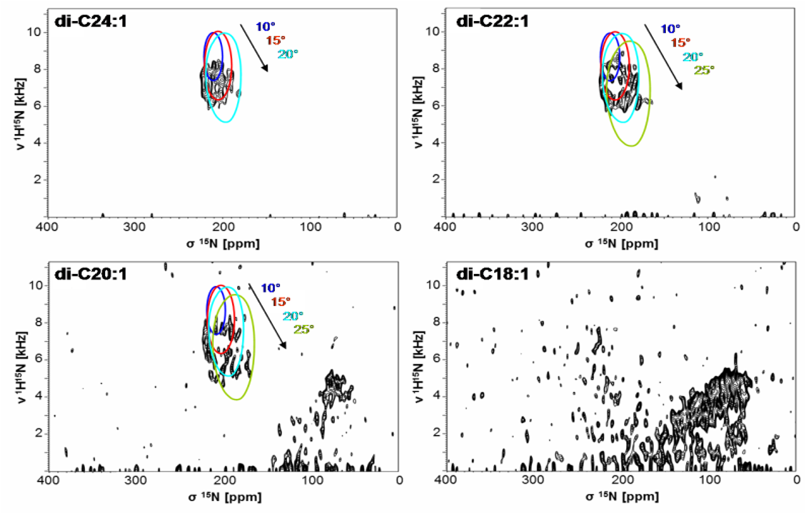
The properties of the lipid bilayer are fundamentally important in controlling the interactions with small peptides, and they are also known to regulate the functional states of larger membrane proteins. For example, solid-state NMR analysis of (conventionally 15N-labeled) transmembrane segments from tyrosine kinase receptors and viral oncoproteins showed that their stability and dimerization behavior is determined critically by the membrane thickness [14], as illustrated by PISEMA-type spectra. The underlying concept of hydrophobic mismatch has been further extended to amphiphilic model peptides, which we could utilize as molecular rulers to measure bilayer thickness in different types of living cells. As a general rule for antimicrobial peptides, however, we found that it is primarily the spontaneous lipid curvature that determines their ability to insert into the hydrophobic bilayer core [15, 16].
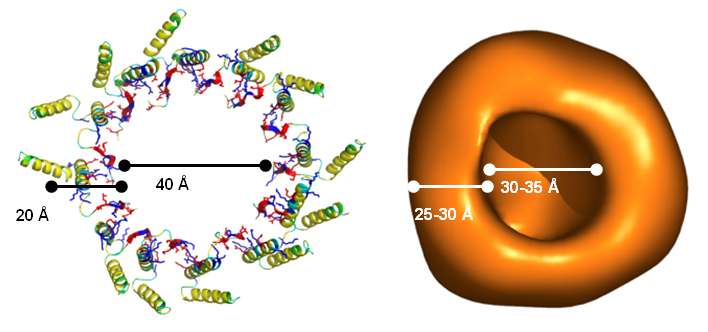
Protein secretion across cellular membranes requires complex machineries. We are studying the bacterial Twin-arginine-translocase (Tat), which is able to export fully folded proteins in bacteria and chloroplasts, driven by pH and the transmembrane potential. Based on the monomeric NMR structure of its pore-forming subunit TatA in membranes [17], we discovered a unique mechanism of folding and self-assembly into a pore with variable size that can adjust to the diameter of the cargo [12]. Oligomerization of TatA is accomplished by ladders of salt bridges between amphiphilic segments, just like in the TisB peptide above. This “charge zipper” concept thus represents a novel structural motif that is predictaed to occur also in other membrane proteins that are currently being explored. Our working hypothesis on the full Tat translocation mechanism is supported by initial NMR and CD data on the TatC receptor [18] and the signal peptide [19]. AFM, FRET and FCS experiments are used to monitor the reconstitution of functional TatA/TatC/SP complexes in giant lipid vesicles, to operate as a pH-driven nanomachine.
Selected publications:
1. Tkachenko, A.N., D.S. Radchenko, P.K. Mykhailiuk, S. Afonin, A.S. Ulrich, I.V. Komarov (2013) Angew. Chem. Int. Ed. 52, 6504-6507
2. Tkachenko, A.N., P.K. Mykhailiuk, S. Afonin, D.S. Radchenko, V.S. Kubyshkin, A.S. Ulrich, I.V. Komarov (2013) Angew. Chem. Int. Ed. 52, 1486–1489
3. Mykhailiuk, P.K., S. Afonin, G.V. Palamarchuk, O.V. Shishkin, A.S. Ulrich, I.V. Komarov (2008), Angewandte Chem. Intl. Ed. 47, 5765-5767
4. Ieronimo, M., S. Afonin, K. Koch, M. Berditsch, P. Wadhwani, A.S. Ulrich (2010) J. Am Chem. Soc. 132, 8822-8824
5. Afonin, S.E., S.L. Grage, M. Ieronimo, P. Wadhwani, A.S. Ulrich (2008) J. Am. Chem. Soc. 130, 16512
6. Wadhwani, P., J. Buerck, E. Strandberg, C. Mink, S. Afonin, A.S. Ulrich (2008) J. Am. Chem. Soc. 130, 16515-16517
7. Maisch, D., P. Wadhwani, S. Afonin, C. Böttcher, B. Koksch, A.S. Ulrich (2009) J. Am. Chem. Soc. 131, 15596–15597
8. Wadhwani, P., E. Strandberg, N. Heidenreich, J. Bürck, S. Fanghänel, A.S. Ulrich (2012) J. Am. Chem. Soc. 134, 6512–6515
9. Berditsch, M., S. Afonin, T. Vladimirova, P. Wadhwani, A.S. Ulrich (2012) Front. Immunol. 3, 222
10. Babii, O., S. Afonin, M. Berditsch, S. Reißer, P.K. Mykhailiuk, V.S. Kubyshkin, T. Steinbrecher, A.S. Ulrich, I.K. Komarov (2014) Angew. Chem. Int. Ed. 53, 3392–3395
11. Steinbrecher, T., S. Prock, J. Reichert, P. Wadhwani, B. Zimpfer, J. Bürck, M. Berditsch, M. Elstner, A.S. Ulrich (2012) Biophys. J. 103, 1460-1469
12. Walther, T.H., C. Gottselig, S.L. Grage, M. Wolf, A.V. Vargiu, M.J. Klein, S. Vollmer, S. Prock, M. Hartmann, S. Afonin, E. Stockwald, H. Heinzmann, O.V. Nolandt, W. Wenzel, P. Ruggerone, A.S. Ulrich (2013) Cell 152, 316–326
13. Walther, T.H., A.S. Ulrich (2014) Curr. Opinin. Struct. Biol. 27, 63–68
14. Muhle-Goll, C., S. Hoffmann, S. Afonin, S.L. Grage, A.A. Polyanski, D. Windisch, M. Zeitler, J. Bürck, A.S. Ulrich (2012) J. Biol. Chem. 287, 26178-26186
15. Strandberg E., D. Tiltak, S. Ehni, P. Wadhwani, A.S. Ulrich (2012) Biochim. Biophys. Acta- Biomembranes 1818, 1764–1776
16. Strandberg, E., J. Zerweck, P. Wadhwani, A.S. Ulrich (2013) Biophys. J. 104, L9–L11
17. Walther T.H., S.L. Grage, N. Roth, A.S. Ulrich (2010) J. Am. Chem. Soc. 132, 15945-15956
18. Nolandt, O.V., T.H. Walther, S. Roth, J. Bürck, A.S. Ulrich (2009) Biochim. Biophys. Acta - Biomembranes 1788, 2238-2244
19. Klein, M.J., S.L. Grage, C. Muhle-Goll, J. Bürck, S. Afonin, A.S. Ulrich (2012) Biochim. Biophys. Acta - Biomembranes 1818, 3025-3031